As deforestation progresses in the tropics, wildlife populations are increasingly restricted to forest fragments. Here we study genetic population structure in the endangered Ashy red colobus (Piliocolobus tephrosceles) population in the forest fragments surrounding Kibale National Park, Uganda. Subsequently, we use landscape features (elevation, road data and distance to the park) to design a feasible strategy to restore forest in a fashion suitable for both the dispersal patterns of the species and land use practices of the local people. A lack of association between geographic distance and pairwise genetic relatedness among localities, the presence of first degree relatives across localities, and a low global Fst value suggest that red colobus individuals have migrated across this landscape in the recent past. Thus, a series of “stepping stone” forests from the fragments to the park will likely maintain viability of red colobus fragment populations. In this area, low-lying valleys are legally protected to prevent flooding and are considered of low-economic value to local people. We identify such valleys for development of community-based forest restoration efforts that will aid in red colobus conservation and provide various ecosystem services. Our study outlines how genetics and community-based restoration can be integrated to provide realistic conservation solutions.
Habitat loss and degradation are major drivers of terrestrial biodiversity declines (Wilson et al., 2016). Globally, approximately 60 million hectares (ha) of tropical old-growth forest were lost between 2002 and 2019 (Weisse and Gladman, 2020). To put this in perspective, an area of tropical forest larger than Madagascar was lost in 18 years, and the rate of loss is accelerating with the amount of tropical forest loss increasing by 2101 km2 each year (Hansen et al., 2013). Today, large areas of intact forest are rare. They comprise only 20% of remaining tropical forest, only 12% of these areas are protected, and these forests are disappearing at a rate of 7.2% annually (Potapov et al., 2017). Furthermore, it is predicted that in the next 50 years there will be a 33-fold increase in the number of fragments (Taubert et al., 2018). As a consequence, many animal species are now only found in fragmented landscapes.
Habitat fragmentation is a landscape-scale process in which a mostly continuous habitat is broken apart into smaller pieces (fragments) scattered within a matrix of less suitable habitat. This process results in the loss of habitat and its subdivision (fragmentation) into a variable number of fragments (Fahrig, 2003, 2017). For the forest-dependent animals in these landscapes, not only are the size and quality of the remaining forest habitats essential, but the nature of the landscape between the remaining forest – the matrix – can also be critical (Galán-Acedo et al., 2021). The matrix can offer resources and allow animals to move through it, or it can act as a barrier, reducing movement and dispersal which can result in isolated populations in which individuals have limited breeding opportunities (Niebuhr et al., 2015). As genetic divergence increases among populations, inbreeding and genetic drift increase within populations, ultimately leading to genomic erosion, loss of adaptive potential, loss of fitness, mutational meltdown, and potentially extinction. For species experiencing such effects, restoring and reconnecting forested habitat may be the only way to ensure their long-term survival (Arroyo-Rodríguez et al., 2020). However, developing realistic solutions to restore habitat connectivity is challenging. This is because the intervening landscapes among habitat fragments are typically anthropogenically modified and tied to human land use practices, land use rights, and livelihoods. Thus, conservation solutions to habitat fragmentation will more often than not require knowledge regarding animal populations, plant communities, and relationships between humans and nature.
In this study, we provide a framework for developing a spatially explicit conservation solution that integrates concepts from conservation genetics and restoration ecology to reverse the effects of habitat fragmentation on an endangered species while providing benefits to the livelihoods of local landowners. We studied the Ashy red colobus monkey (Piliocolobus tephrosceles) population living in Kibale National Park (Uganda) and surrounding forest fragments. The Ashy red colobus is an endangered species threatened primarily by habitat loss and fragmentation (Linder et al., 2021). It is a long-lived arboreal folivore and forest dependent species with social groups occupying an annual home range of between 35 and 71 ha (Struhsaker, 1980). These monkeys have limited ability to use and move through non-forested habitats, because they never use swamps, and rarely raid crops or descend to the ground (Chapman unpublished data). The red colobus population inside the park boundaries has been the focus of >50 years of research, it is the world’s largest known population of Ashy red colobus, with an estimated size of 30,000 individuals and is growing at an annual rate of 3% (Chapman et al., 2018). In contrast, between 2000 and 2010, the red colobus populations in the surrounding areas declined by 83% (Chapman et al., 2013b). Thus, these populations outside of the park have a bleak outlook unless concerted efforts are made to mitigate the effects of habitat loss and fragmentation. We applied population genetics to infer patterns of migration among forest fragments in the Ashy red colobus. Then, we used site specific landscape features, information on community values and land use, and knowledge of land quality and protection status, to identify spatially explicit areas between fragments and the main forest where forest restoration is feasible. We subsequently intersected these two results to determine which fragment populations should be prioritized to reconnect to the main forest through a community-based forest restoration strategy that will improve viability of certain red colobus fragment populations and simultaneously provide ecosystem services to the local community.
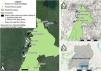
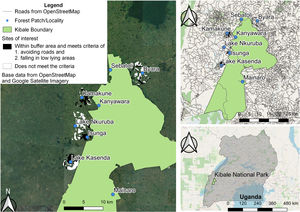
Material and methodsStudy site and samplingThis study was conducted in Kibale National Park, Uganda (Fig. 1), a 795 km2 protected area in western Uganda (0° 13′–0° 41′N and 30° 19′–30° 32′E) (Chapman et al., 2018). The process of forest fragmentation outside of Kibale was well advanced by the 1940s, and aerial photographs of the region taken in 1959 reveal only small fragments embedded in a matrix of agricultural fields. Improvement of the roads to the region in the early 1990s and a rapidly growing human population density led to rapid forest clearing (Chapman et al., 2013b). Between 2000 and 2020 the human population density within 1 km of the park’s boundary almost doubled, from 123 to 229 people/km2 (MacKenzie et al., 2017). Almost all (95%) of the local people are smallhold farmers, cultivating less than 5 ha (Mackenzie and Hartter, 2013) and woodlots are a profitable source of income for many families (Naughton-Treves et al., 2007). As a result, the forest fragments, that comprise 25% of the landscape, are now isolated and imbedded in a landscape of small-scale farms often with associated woodlots (46%), and swamps (29%; Hartter et al., 2015).
Right: map of the study area in Western Uganda showing Kibale National Park and the sampled areas with red colobus presence both within the park and in forest fragments outside the protected park. Left: close up of the study area where the areas in black neighboring the fragments are lowland areas that would be the most likely restoration areas to plant trees to act as forested stepping stones connecting the fragments to Kibale.
Between 2010 and 2013 we collected 299 fecal and blood samples from eight sites in or near Kibale National Park. We sampled three sites within Kibale (Kanyawara, Sebitoli, and Mainaro) and five fragments (Isunga, Kamakune, Byara, Lake Nkuruba, and Lake Kasenda). The characteristics of the fragments vary depending on location, protection status, size, and distance to the park (straight line distance ranges from 0.96 to 3.42 km; Fig. 1), but they are all in areas difficult to farm such as swamps and steep slopes associated with crater lakes (Hartter et al., 2011).
Population genetic analysesSamples were genotyped at 15 microsatellite loci used in previous studies of colobus monkeys (raw data available at http://hdl.handle.net/10261/261482, see Supplementary for details). We tested for deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium for each sampled location and overall population in GENEPOP version 4.0.10 (Rousset, 2008). We assessed the levels of genetic diversity by calculating the mean unbiased expected and observed heterozygosity and mean allelic richness (GENALEX 6, Peakall and Smouse, 2006). We calculated the mean allelic richness and the number of private alleles adjusted for sample size using rarefaction in HP-RARE 1.0 (Kalinowski, 2005). We used Fstat v. 2.9.3.2 (Goudet, 1995) to calculate and test the significance of both FIS values for each site and across sites (Weir and Cockerham, 1984) and overall and pairwise FST values across sites (10,000 permutations). We inferred population structure using STRUCTURE v. 2.3.4 (Pritchard et al., 2000). Using an admixture model with LOCPRIOR we tested K = 1–6 with 10 replicates for each K-level, with an initial burn-in of 2.5 × 105 followed by 7.5 × 105 Monte Carlo Markov Chain iterations. The most likely number of clusters was identified using posterior probability (Pritchard et al., 2000). To avoid bias because of the presence of related individuals, we generated a subset of individuals which did not include first degree associations (full-sibling and parent-offspring) identified through MLrelate (Kalinowski et al., 2006). We reran both Fstat and STRUCTURE using this subset. We also calculated pairwise genetic relatedness, mean genetic relatedness for each site, pairwise mean genetic relatedness between sites, and calculated the proportion of first-degree relationships shared between different sites. Using the package adegenet 2.1.0 (Jombart and Ahmed, 2011) in R (R-Core-Team, 2020) we ran Mantel tests to test for isolation by distance and analyzed if mean relatedness and proportion of first-degree relationships decreased significantly with distance (see Supplementary for details).
Spatial analysisFor the spatial analysis we focused on connecting fragments to the large population of red colobus in the park. We did not consider connecting fragments together as the distance among them is large and the small number of red colobus individuals per fragment (average = 17 animals; Chapman et al., 2013b) does not allow for a viable fragment meta-population to persist in isolation from the park. The landscape outside the national park and surrounding the fragments is heavily modified agricultural land. However, wet valley bottoms are less desirable areas, as establishing agriculture is difficult and less profitable. Furthermore, wetlands are officially government owned and under National Environment Regulations (for wetlands, riverbanks, and lake shore management), and the land immediately adjacent to these wet areas cannot be legally degraded (Isunju and Kemp, 2016). Thus, legislation and perceived low value makes restoring wet valley bottoms and the immediately surrounding areas more feasible than restoring other types of land cover.
To identify valley bottoms, between fragments and the park, we used 30 m elevation data from The Shuttle Radar Topography Mission (Farr et al., 2007) and roads data from OpenStreetMap. The threshold for low-lying areas for each fragment was determined by examining the elevation profile between the patch and the park. Areas likely representing wetlands were those exhibiting the same elevation along a typical dendritic path and were bordered by areas of increasing elevation. These areas were confirmed as being wetland by examining topographic maps, satellite images, and ground surveys. The proposed forest restoration areas are between the park and 3.67 km (the farthest location of existing patches) and within a radius, where the radius is determined by the shortest distance between the park and the particular fragment. Since most settlements occur close to roads, a 200 m buffer was applied around roads to account for existing villages and to leave space for future development and this area was excluded from further consideration as a corridor. All spatial analyses were carried our using QGIS (QGIS.org, 2022).
ResultsGenetic diversity, structure, and relatednessWe successfully genotyped 234 samples and identified 159 individuals (Table 1). All loci analyzed were polymorphic at the 8 sampled sites and did not deviate significantly from expected genotype frequencies under HWE, showed evidence of linkage disequilibrium or evidence of null alleles. None of the sites nor the overall population deviated significantly from HWE. Measures of genetic diversity were similar across all sites (see Table 1 for summary statistics of genetic diversity). Overall FST was 0.028 (Upper/lower 95% CI = 0.010−0.045) and was significantly different from zero (p-value < 0.002). When pairwise FST were carried out across sites, 4 pairwise FST values were significant for one site, Nkuruba (Table 2). However, when we removed first-degree related individuals within localities from the analyses it was no longer significant. STRUCTURE analyses showed that all sites belong to a single cluster because lnP(D) = −5598.9 was maximized at K = 1. We found that there is not a significant relationship between genetic distance and geographic distance among fragments and main forest. Mean pairwise genetic relatedness per site ranged from 0.077 in Kanyawara to 0.190 in Nkuruba (mean across sites +/− standard deviation = 0.124 ± 0.035), and was higher in the fragments than in the main forest (mean sites +/− standard deviation: Forest fragments = 0.129 ± 0.033, Main forest = 0.104 ± 0.047; Table 2 Supplementary material). Pairwise relatedness between fragments, as well as proportion of first-degree relationships, was not significantly associated with distance among sampled localities.
Genetic diversity measures for the locations included in the analyses. N is the sample size per fragment; Na is the number of alleles; Ne is the number of effective alleles (number of alleles with frequency > 5%); Ac is the allelic richness corrected by sample size; Pc is the proportion of private alleles corrected by sample size; Ho is observe heterozygosity; He is the expected heterozygosity and UHe is the unbiased expected heterozygosity; FIS is the inbreeding coefficient; and σ is the standard deviation.
| Pop | N | Na | Ne | Ac | Pc | Ho | He | UHe | FIS |
|---|---|---|---|---|---|---|---|---|---|
| Kanyawara | 77 | 4.933 | 3.097 | 3.620 | 0.040 | 0.627 | 0.621 | 0.625 | −0.003 |
| Sebitoli | 19 | 4.400 | 2.876 | 3.590 | 0.020 | 0.606 | 0.588 | 0.605 | −0.001 |
| Mainaro | 10 | 3.467 | 2.564 | 3.260 | 0.100 | 0.547 | 0.559 | 0.595 | 0.086 |
| Isunga | 11 | 3.867 | 2.967 | 3.520 | 0.010 | 0.573 | 0.614 | 0.646 | 0.119 |
| Kamakune | 9 | 4.000 | 2.702 | 3.610 | 0.100 | 0.663 | 0.588 | 0.623 | −0.068 |
| Byara | 9 | 3.667 | 2.665 | 3.440 | 0.010 | 0.645 | 0.578 | 0.617 | −0.048 |
| Lake Nkuruba | 16 | 3.867 | 2.525 | 3.300 | 0.010 | 0.650 | 0.576 | 0.596 | −0.094 |
| Lake Kasenda | 8 | 3.667 | 2.675 | 3.460 | 0.060 | 0.571 | 0.568 | 0.608 | 0.063 |
| Total/Mean | 159 | 3.983 | 2.759 | 3.475 | 0.044 | 0.610 | 0.587 | 0.614 | 0.007 |
| σ | 0.443 | 0.188 | 0.129 | 0.036 | 0.040 | 0.020 | 0.016 | 0.072 |
Pairwise FST between sampled sites calculated including all the individuals are on the lower half. Euclidean Distances (km) between sampled sites are on the upper half. Significant pairwise comparisons are in bold text (p < 0.002).
| Kanyawara | Sebitoli | Mainaro | Isunga | Kamakune | Byara | Nkuruba | Kasenda | |
|---|---|---|---|---|---|---|---|---|
| Kanyawara | 0 | 9.582 | 23.038 | 8.613 | 4.788 | 13.950 | 7.948 | 16.120 |
| Sebitoli | 0.009 | 0 | 31.869 | 18.193 | 10.159 | 8.279 | 16.896 | 25.635 |
| Mainaro | 0.024 | 0.054 | 0 | 16.052 | 26.038 | 31.972 | 20.011 | 13.656 |
| Isunga | 0.012 | 0.020 | 0.014 | 0 | 11.237 | 21.658 | 4.219 | 7.629 |
| Kamakune | 0.022 | 0.028 | 0.053 | 0.033 | 0 | 16.752 | 7.441 | 16.713 |
| Byara | 0.006 | 0.016 | 0.017 | 0.010 | 0.021 | 0 | 21.883 | 29.248 |
| Nkuruba | 0.044 | 0.056 | 0.095 | 0.039 | 0.074 | 0.063 | 0 | 9.292 |
| Kasenda | 0.028 | 0.034 | 0.056 | 0.017 | 0.050 | 0.048 | 0.055 | 0 |
There are many areas of wet valley bottoms between the forests of Kibale and the fragments, providing opportunities to construct forested stepping stones, i.e. unconnected areas of semi-natural habitat that would allow Ashy red colobus individuals to move between fragments (Fig. 1). The shortest distance between the edge of the forest fragments and the park averaged 2292 m (range: 1007–3675 m; Table 3). Isunga (1007 m) was the fragment closest to the national park, followed by Kasenda (2072 m), Kamakune (2522 m), Byara (3140 m), and Nkuruba (3675 m). The percentage of this distance that was not along valley bottom averaged 66% (range: 17–86%; Table 3). The distance not along valley bottom averaged 1562 m (range: 352–3161 m; Table 3). Ultimately, the percent of land meeting the favorable criteria in the different locations (i.e. were along wet valley bottoms) was: Nkuruba (14%), Byara (17%), Isunga (23%), Kamakune (41%) and Kasenda (83%).
The distance between the forests of Kibale National Park, Uganda and the fragments and the proportion of this distance that is not valley bottom (i.e it does not meet the criteria for restoration).
| Name | Shortest distance to Park (m) | Distance not in valley bottom habitat (m) | % of distance not in valley bottom habitat |
|---|---|---|---|
| Byara | 3140 | 2606 | 83% |
| Kamakune | 2522 | 1488 | 59% |
| Nkuruba | 3675 | 3161 | 86% |
| Isunga | 1007 | 776 | 77% |
| Kasenda | 2072 | 352 | 17% |
Tropical forests are becoming increasingly fragmented bringing hundred of species to the brink of extinction. Conservation genetics helps to asses how habitat fragmentation affects species. Restoration ecology, which considers cultural values in restoring plant communities, is well suited to reversing the effects of habitat fragmentation. However, these two disciplines are often not integrated into a single framework. This is necessary as the effects of habitat fragmentation vary across systems, and matrix habitats between tropical forests are typically maintained by various human activities tied to livelihoods and needs. Thus, mitigating the effects of tropical forest fragmentation and slowing future biodiversity declines will rely on an integration of expertise from disparate disciplines. Here, we demonstrate an application of this framework on the endangered Ashy red colobus monkey surrounding Kibale National Park in Uganda.
Recent red colobus dispersal among fragmentsOur genetic results suggest that gene flow is either occurring or has recently occurred at the landscape level in this red colobus population. This is evidenced by the presence of little genetic differentiation among localities (global FST value lower than 0.03), no pairwise FST value among localities significantly different from zero after controlling for related individuals within localities, only one cluster detected in the structure analysis, and the presence of first-degree relatives among different fragments, which indicates recent movement of individuals among localities. Lastly, we found that genetic diversity is similar across localities. While it is sometimes difficult to detect the genetic effects of habitat fragmentation, it is often because a species is resilient to habitat disturbance and/or not enough generations have passed for differences in allele frequencies to accrue among fragmented populations (Landguth et al., 2010). However, previous studies of red colobus response to habitat fragmentation indicate that they rarely move among fragments and show high fragment fidelity by remaining in a slowly degrading fragment until only a dozen or so trees are left (Chapman et al., 2007). Furthermore, fragmentation in the Kibale area was established by the 1940s. Thus, while red colobus have a relatively long generation time (around 10 years), a sufficient amount of time has passed for the genetic effects of habitat fragmentation to be detected. It thus appears that red colobus individuals have been able to either disperse through the matrix in-between fragments, use the matrix effectively, and/or use suboptimal and/or agricultural habitat, such as Eucalyptus tree plantations (Chapman et al., 2013a).
Despite these results, we caution that red colobus use of matrix habitats is unlikely to be viable in the long-term as fragments continue to degrade and become further apart, and it is possible that the evidence of dispersal found in this study is from one or two generations ago (10–20 years prior to sampling in 2010–2013). Also, the red colobus in the fragments generally have high average genetic relatedness, likely because the fragments are small and most individuals within them belong to a single social group, thus increasing the likelihood of inbreeding if individuals cannot disperse. Regardless, we find these results encouraging, in that connectivity was present in the recent past. This result contrasts with other studies of red colobus fragmentation that have shown a clear effect on genetic population structure (Minhós et al., 2016; Ruiz-Lopez et al., 2016) and suggests that each habitat fragmentation scenario should be considered separately prior to design of conservation interventions. Given our evidence that dispersal has recently occurred across this landscape, a system of forested stepping stones is likely sufficient to encourage red colobus dispersal from the fragment populations to the park into the future, thus improving the conservation value of these forest fragments.
Placement and design of reforested islandsDeveloping a community-based forest restoration effort concentrated in and around the valley bottoms could provide an effective conservation intervention that considers multiple stakeholder groups. Such an approach is necessary because land purchase is financially prohibitive and inappropriate given the history of land conflict in the area (MacKenzie et al., 2011). Our spatial analysis suggests that establishment of simple forested stepping stones between the study fragments and the park is more likely to succeed in certain areas. While other socioeconomic, cultural, and logistic factors need to be evaluated, the genetic and spatial information produced here specifically point to Kasenda and Isunga as the fragments where restoration of forests within and surrounding valley bottoms is most likely to succeed in enhancing red colobus conservation. Kasenda forest fragment contains the largest percentage of low-lying habitat between itself and the park, and it contains a small ecotourism facility surrounding a crater lake, thus providing protection from agricultural conversion as well as incentives to maintain a healthy wildlife community. Meanwhile, the Isunga fragment is substantially closer to the park compared to the other fragments.
Growing trees for fuelwood has become one of the most profitable cash crops in the study region. Wood supplies over 80% of domestic energy needs across Africa, including 88% in Uganda and 95% in the Democratic Republic of Congo (Mayaux et al., 2013), and many urban dwellers in Uganda still heavily rely on fuelwood for cooking. Thus, if local people could be encouraged to plant woodlots comprised of native species in the proposed forest restoration areas along valley bottoms, these could serve as the stepping stones for red colobus dispersal. For these woodlots to be sustainable in the long-term, farmers might have to harvest trees asynchronously. However, as different sizes of trees can be cut and sold to different markets (fuelwood, poles for building, large trees for timber), it is not unreasonable to expect that many woodlots could be harvested in a way to always maintain their stepping stone function. As many of the hardwoods that grow in the region now secure high prices for furniture, it should also be possible to encourage the planting of native tree species that would provide red colobus food as they pass through (e.g., Markhamia lutea). Detailed study on the effects of incorporating non-native tree species preferred by local farmers, such as Eucalyptus, or isolated trees should also be conducted. Lastly, careful consideration of the costs and benefits of a community-based forest restoration project is needed, especially compared to alternative conservation projects, as reforestation can be expensive. However, there are already over 70 registered climate mitigation projects to reduce emissions from deforestation and degradation, some of them protecting areas that are similar to Kibale from an ecological and socioeconomic perspective (e.g. Dickson et al., 2020). For example, the Ntakata community project involves 8 villages, with more than 36,000 people and has prevented the cut of 1,250,000 trees. Thus, we believe it is possible that funding for the proposed community-based forest restoration could come from carbon offset programs, as such a project would benefit local and global human communities (see Box 1 for more information).
A community-based forest restoration project — what are the costs and benefits?
A community conservation program such as we envision would be a substantial financial investment and require long-term conservation commitment. Based on over 30 years of running similar projects in the region, we estimate it would cost a minimum of $80,000–$100,000 a year (excluding foreign supervisors’ salary or permanent equipment). To have the required impact (i.e., to encourage the planting, growth, and maintenance of sufficient forested stepping stones), the project would need to run for ten years, preferably longer. Thus, a ten-year project would likely cost a million dollars, and if foreign salaries are included the price would only increase. Thus, a careful assessment of the benefits of such an investment relative to other conservation initiatives is needed, and an important consideration should be the source of the conservation funds and competing conservation needs. If such a community conservation project were only for the benefit of a couple of red colobus fragment populations outside of Kibale, then this cost is likely too high. However, given that protecting and restorating native ecosystems grew out of the 1997 Kyoto protocol as a major strategy for mitigating and adapting to climate change, funds for this effort could potentially come from the carbon-offset market and thus not be in competition with resources for Kibale National Park management and Ashy red colobus projects prioritized in the IUCN Red Colobus Conservation Action Plan (Linder et al., 2021). High-income, high-carbon emitting countries are willing to support projects that sequester carbon by growing trees (Wheeler et al., 2016). These projects involve large sums of money, as $300 million of credits had been sold on voluntary markets by 2017; however, Africa accounted for just $20 million of this global total (UNEP, 2019) and the continent is seen as a promising market. Thus, development of a community-based forest restoration project could have benefits for multiple stakeholders with a potential end result of increasing the conservation value of forest fragments for the Ashy red colobus, complementing the important conservation efforts occurring within Kibale National Park, improving the livelihoods of local farmers, and mitigating the global and local effects of climate change.
In summary, our study shows how genetic information can be used to inform a spatially explicit strategy of community-based forest restoration. This type of community-based conservation project would require substantial long-term funding and commitment as well as cost-benefit assessment, but it could provide an additional conservation tool for the endangered Ashy red colobus. Funding for such an effort could potentially come from carbon offset projects and thus not compete for funding for other conservation activities. We encourage an increased integration between conservation genetics and restoration ecology for the development of realistic solutions for wildlife conservation and management.
Authorship contribution statementMJR, NT, CAC, and TLG designed the study, TLG, CAC, and NT obtained the funding, MJR, CAC, NDS, NT, PO, AJH, TLG, and DS, collected the data, MJR, AJH, and DS analyzed the data, MJR, NT, DS, and CAC wrote the paper, and all authors provided feedback, comments, and edits on the writing and approved the final submission.
Declaration of competing interestWe declare that there are no known competing financial interests or personal relationships that could have influenced this study.
We thank the Uganda Wildlife Authority and Uganda National Council of Science and Technology for permission to conduct this research. We are grateful to Robert Basaija, Peter Tuhairwe, Clovice Kaganzi, and Dr. Dennis Twinomugisha for assistance with logistics and fieldwork. Financial support for this research was provided by NIH grant TW009237 as part of the joint NIH–NSF Ecology of Infectious Disease program and the UK Economic and Social Research Council, NSF BCS-1540459, National Geographic Society, Leakey Foundation, NSERC, the IDRC grant “Climate change and increasing human-wildlife conflict”, and the University of Oregon.