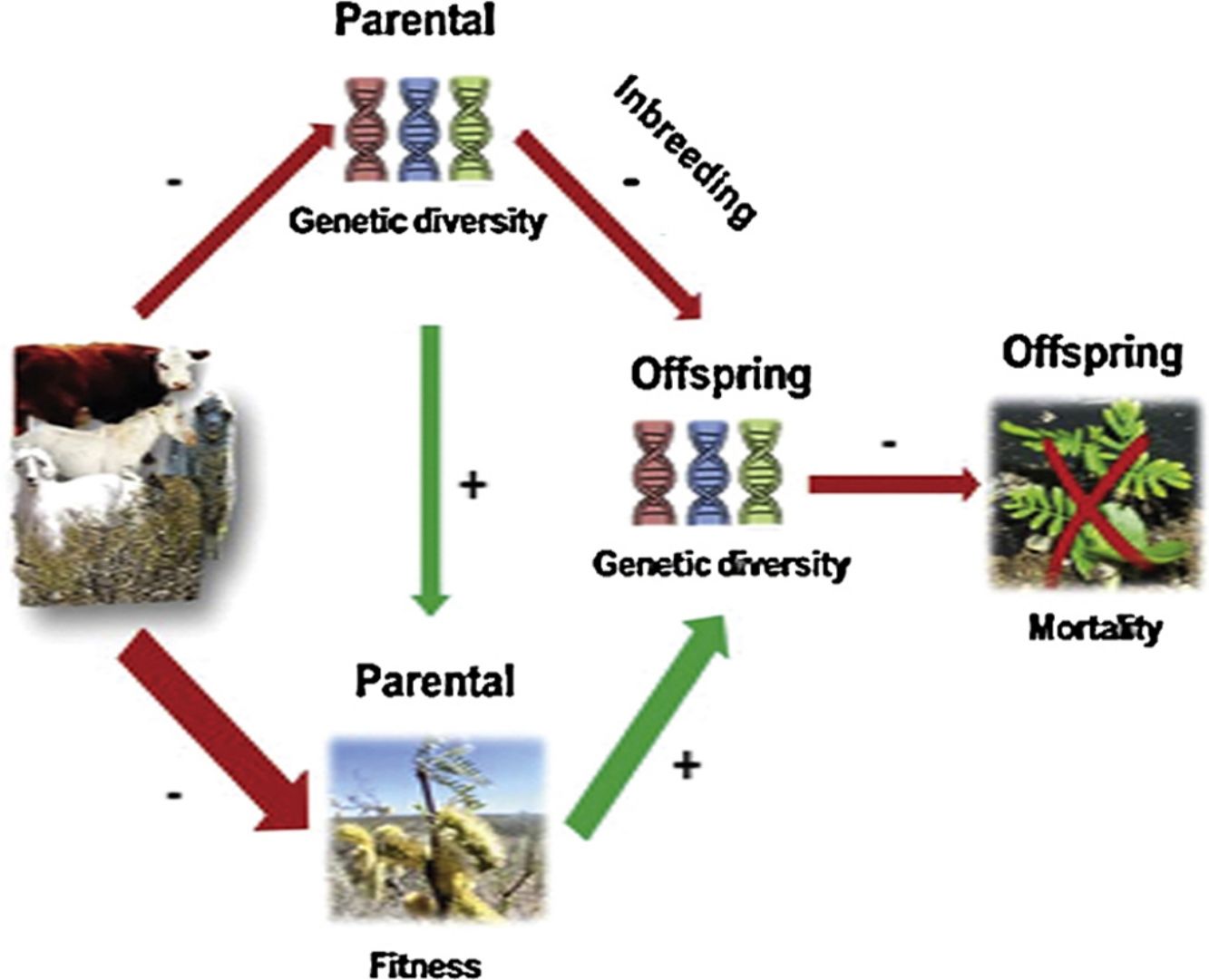
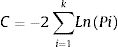Biodiversity conservation focuses on species and/or populations, but preserving genetic diversity and structure has received limited attention and even less maintaining species evolutionary potential over generations. Genetic diversity is an essential component of biodiversity enabling species’ persistence, particularly under disturbances. Via sexual reproduction genetic diversity is transmitted across generations and greater outcrossing in parental populations will lead to greater genetic diversity in their offspring. Grazing by exotic large herbivores is one of the main disturbances driving biodiversity loss threatening rangelands sustainability worldwide. We investigated grazing effects on fitness and genetic diversity of parental and offspring cohorts of Prosopis alpataco from Patagonian Monte Desert. We collected fresh leaves and seeds from 10 independent rangelands with different herbivore density, forming a grazing gradient, and estimated genetic parameters from allele frequencies using isoenzymes. We recorded plant size, seed weight, seed set, seedling emergence and mortality as proxies of plant fitness. Applying regression models and path analysis (D-separation) approaches we observed that increasing grazing reduced seed set and seedling emergence, and significantly increased seedling mortality. Parental and offspring suffered from inbreeding. Moreover, we found genetic diversity loss throughout cohorts in all rangelands, however, this loss was relatively lower at intermediate grazing intensities. The introduction of large herbivores in unmanaged rangelands affected vegetation structure, jeopardizing their evolutionary potential and system sustainability. Therefore, natural revegetation may be compromised by aggravated genetic diversity losses along generations that might be deepened in drylands under forecasted climate change. This highlights the importance of evaluating and conserving genetic diversity.
Genetic diversity is essential for sustaining biodiversity over time given that a greater genetic diversity increases species likelihood to respond to environmental changes and disturbances (Banks et al., 2013; Hoffmann and Parsons, 1991; Rice and Jain, 2013). Land transformation and fragmentation caused by livestock breeding is affecting ecosystems worldwide (FAO, 2019), although for some areas are reported positive effects (García-Fernández et al., 2019; Shapira et al., 2019), while for others negative effects were observed (Fischer and Lindenmayer, 2007; Hobbs et al., 2008; Yirdaw et al., 2017). Particularly, it is well documented that livestock grazing in arid ecosystems leads to severe reduction in vegetation cover, abundance and richness, affecting their functioning (Li et al., 2017; Paruelo and Aguiar, 2003; Tadey, 2006). Moreover, long-term grazing and its negative impact on ecosystem dynamics can have unpredictable evolutionary consequences on plant communities (Hanke et al., 2014; Herman and Sultan, 2011; Tadey and Farji-Brener, 2007). However, the extent of these effects on plant genetic diversity has received limited attention, and even less the effect of grazing on genetic diversity through generations (Fu et al., 2005; Wu et al., 2010).
Large herbivores can directly or indirectly modify plant fecundity and gene flow during pre-dispersal and post-dispersal stages, which might lead to changes in genetic diversity and structure of consumed plant populations (Grant, 2010; Mulder, 1999). Even if herbivores can help plant species dispersal by zoochory (Bessega et al., 2017; Campos et al., 2011; García-Fernández et al., 2019), reductions in effective population size by consumption of individuals (either by killing or damage) and/or decreasing reproduction (by consumption of flowers and/or fruits) might result in less breeding individuals contributing to the next generation. This process may lead to the reduction of the species’ gene pool and the restriction of gene flow, eroding over time, genetic diversity and increasing divergence among populations (Couvet, 2002; Lanfear et al., 2014; Leimu et al., 2006; Young et al., 1996). Small populations are subjected to inbreeding effects as a consequence of the increased likelihood of mating with closely related individuals (Heinken and Weber, 2013). However, inbreeding may also be increased through genetic drift, changes in allele frequencies caused by cumulative effects and bottleneck, which is a kind of genetic drift where the gene pool of a population is drastically reduced by a stochastic event (Luikart and Cornuet, 1998; Luikart et al., 1998; Piry et al., 1999). Usually, after these events inbreeding may be deepened by limited reproduction, decreasing offspring viability and fitness (i.e. inbreeding depression) (Hedrick and Kalinowski, 2000). Consequently, reductions in genetic variation could severely decrease the adaptive capacity of plant populations, affecting their ability to cope with future natural and/or anthropogenic impacts in a changing world (Cornuet and Luikart, 1996; Frankham et al., 2015; Luikart and Cornuet, 1998; Luikart et al., 1998; Williams, 2001).
From an ecological perspective, a limited number of available mates as a consequence of smaller population size may also affect future generations through reduced pollination and reproduction (Ågren, 1996). The number of mates (i.e. flowering plants) may affect pollination attraction and foraging behavior (Lehtilä and Strauss, 1997; Tadey, 2015, 2008), leading to lower pollen flow within population (Lehtilä and Strauss, 1997). When plants are less attractive, pollinators tend to reduce visitation frequency or to leave to other rich-resources sites (Strauss et al., 1996). Alternatively, when attractive plants are distant from each other, pollinators tend to stay longer within a single plant (Tadey, 2015). These behaviors restrict pollen flow and increase self-pollination, reducing pollen quality (i.e. endogamic pollen) (Aguilar et al., 2008; de Vere et al., 2009; Herrera, 2000; Labouche et al., 2017; Mothershead and Marquis, 2000; Oostermeijer et al., 1994; Tadey, 2008). These last processes may result in inbreeding depression expressed as lower reproduction success by reduced fruit and seed set, germination rates and/or greater seed abortion and seedling mortality (Abrahamsson et al., 2013; de Vere et al., 2009; González-Varo et al., 2012; Oostermeijer et al., 2003; Reed and Frankham, 2003; Waser and Price, 1989). Additionally, plant damage may reduce nutritional resources for new reproductive structures, leading to lower flower production and, consequently, lower plant fitness (Angeloni et al., 2011; Charlesworth and Charlesworth, 1987; Keller and Waller, 2002; Lehtilä and Strauss, 1997; Mothershead and Marquis, 2000; Ramsey and Vaughton, 2016). These negative repercussions of large exotic herbivores on plant fitness may reduce seed banks and seed dispersal, compromising natural revegetation and aggravating genetic diversity losses along generations (Pol et al., 2014; Sternberg et al., 2003; Tadey and Souto, 2016; Tadey, 2007; Trlica and Rittenhouse, 1993).
Particularly in dry environments, large herbivores can be important seed dispersal agents through defecation and trampling, and promoters of seedling establishment for some species by modifying micro-environmental conditions (Aschero and García, 2012; Aschero et al., 2016; Bessega et al., 2017; Campos et al., 2011). In some regions it was observed that livestock may have positive effects on species and ecosystems, such as enhancing seed dispersal and soil quality (Abdalla et al., 2018; Manley et al., 1995;Root-Bernstein et al., 2017; Shapira et al., 2019; Tarin et al., 2016). However, previous works in arid ecosystems showed that livestock may also indirectly affect native dispersers (such as rodents and birds) affecting their interaction with plant species (Marone et al., 2008; Milesi et al., 2002; Tadey, 2019). Additionally, pollen dispersal may also be indirectly affected by livestock grazing. Positive effects were reported for pollination community at intermediate levels of grazing (Tadey, 2015); nevertheless, the increment in pollinators richness and abundance were not reflected in an increase in plant reproduction (Tadey and Souto, 2016; Tadey, 2007). Moreover, a greenhouse study reported increased seedling vigor in some species with increasing livestock density; however, their response is still unknown under natural arid conditions (Tadey and Souto, 2016). Contrarily, other studies found that grazing by large herbivores reduced plant cover, richness, reproduction, germination (Cerda et al., 2012a; Tadey and Farji-Brener, 2007; Tadey, 2006) and even genetic diversity of dominant shrubs (Larrea divaricata and Larrea cuneifolia) (Souto and Tadey, 2018). Therefore, although it seems that livestock may have positive effects in arid regions, finally those effects are counteracted by other negative effects. This underlines the importance of evaluating direct and indirect causal relationships between variables to detect the strongest effect.
Vegetation in the world has evolved in association with herbivory developing adaptive traits. However, in arid regions, such as Patagonian Monte Desert, current vegetation traits suggesting coadaptation are notably weak since browsing reduces reproduction (Cerda et al., 2012b; Tadey and Farji-Brener, 2007; Tadey and Souto, 2016; Tadey, 2020, 2006). This may be partially explained by a weak co-evolution history with herbivores since the large extinct megafauna cited for Monte Desert, or nearby, were mostly described as grazers, found in moister areas associated water and grasses (e.g. records from saline lagoons and endorheic basins), suggesting low association with xeric shrub vegetation (Fernández et al., 2016; Mehl et al., 2019; Tadey, 2020; Villavicencio et al., 2016). In addition, persistent herbivores, such as guanaco (Lama guanicoe, Müller), with a generalist diet including shrubs, are currently scarce in Monte Desert regions and probably were in the past as well (Burgi et al., 2012; Flores et al., 2012). The low abundance of herbivores may also explain a weak coevolution in this xeric vegetation. Desert climatic hostility also modeled vegetation evolution, influencing plant growth which needed to survive under water scarcity and high evapotranspiration rates. Therefore, desert vegetation is characterized by xerophytic lifeforms with slow growth rates and low vegetative reproduction, mainly relying on sexual reproduction to persist over time (Fisher and Turner, 1978). Moreover, as other drylands of the world, Patagonian Monte Desert is an arid ecosystem that is increasingly affected by livestock grazing, which accentuates severe desertification problems, with important socioeconomic impacts on local communities (Maestre et al., 2009; Reynolds et al., 2007). Although it is known that rotation and transhumance practices may mitigate grazing effect (Baied, 1989; Barnes et al., 2008; Root-Bernstein et al., 2017), these grazing management tools are infrequent in Monte Desert (Auffret et al., 2015; García-Fernández et al., 2019; Olea and Mateo-Tomás, 2009). As a consequence of this, serious environmental degradation is observed in most of Patagonian drylands (Ares et al., 2003; INTA, 2011). Estimating the long-term effects of grazing is still a challenge and needs deeper investigation. We studied grazing effects on genetic diversity and fitness in parental and offspring cohorts of Prosopis alpataco Phill., a xerophytic shrub highly consumed by exotic large herbivores in Patagonian Monte Desert. This shrub can be dispersed by livestock, had good germination rates and is abundant in the studied region which makes it suitable to estimate genetic diversity on offspring. To fulfil our aim, we tested the following hypotheses: (1) Plant damage by herbivore consumption of photosynthetic tissue, flowers and fruits reduces the availability of plant resources precluding the allocation to new reproductive structures decreasing plant fitness by reducing the formation of fruits and seeds in terms of quality and quantity. So, as large herbivores increase in abundance, we expect a decrease in plant size (height and diameter), seed weight, seed set and seedling emergence rate; (2) Herbivore consumption of breeding individuals causes lower population reproduction success, eventually decreasing genetic diversity and increasing genetic structure of standing populations, deepening this effect through cohorts. As large herbivores increase in abundance, we expect a decrease in mean number of alleles, effective number of alleles, observed and expected heterozygosity, and percentage of polymorphic loci, and a worsening of these effects in the next cohort. Additionally, we expect that grazing will intensify inbreeding within plant parental populations and that these effects would be deepened in the offspring populations. We assess relative genetic diversity losses between parental and offspring generation (ΔGV) calculated for each genetic parameter separately; expecting greater losses of genetic diversity between generations as grazing intensity increases. Additionally, we tested a hypothetical causal model using D-separation methodology to estimate grazing direct and indirect effects on parental and offspring fitness and genetic diversity. We predicted that individuals with higher genetic diversity will produce heavier seeds, higher seed set and seedling emergence rate, along with lower seedling mortality.
Materials and methodsStudy areaThe study area was located in northwest Patagonia Argentina, in Neuquén province, between Arroyito (39°05′S, 68°35′ W) and Villa "El Chocón" (39°17′ S, 68°55′ W). The vegetation is a xerophytic shrubland that belongs to the Monte Desert phytogeographic region (Cabrera, 1966). This habitat is the largest dry region of South America with a high evaporation rate enhanced by strong westerly winds, low rainfall and high summer temperatures (Abraham et al., 2009; Paruelo et al., 1998; Busso and Fernández, 2017). Mean annual precipitation is below 200 mm and average annual temperature is around 15 °C (Tadey, 2006; Villagra and Roig, 2002), resulting in a strong summer deficit (Leon et al., 1998; Paruelo et al., 1998). Vegetation is scattered and is dominated by Prosopis alpataco and two Larrea species (L. divaricata and L. cuneifolia Cav.), associated with other abundant xerophytic shrub species such as Atriplex lampa Gill. ex Moq, Bougainvillea spinosa (Cavanilles) Heimerl, Monttea aphylla (Miers.) Hauman and Chuquiraga erinacea D. Don (Cabrera, 1966). In this region herbs and grasses represent only 6% of total plant cover (Tadey, 2006). We selected ten rangelands with increasing exotic large herbivore abundance to study its effects on P. alpataco fitness, genetic diversity and structure over generations. All rangelands were located in the same region along Argentine National Route 237, sharing similar environmental conditions, having the same NW orientation and the maximum distance between them was 42 km (Fig. A.1, Appendix A). Rangelands are not fenced, their area varied between 94 and 25,000 Ha and there were no water points within them. Rangelands are operated by poor smallholders, are under continuous grazing throughout the year, have no management (i.e. not subjected to rotation or other practices) and differ in the composition of exotic large herbivores and grazing history (see Appendix A, Table A.1). The abundance of exotic herbivores was provided by rangeland owners. This region is dominated by shrubs, since grasses and herbs are scare, all livestock end up browsing available plant species to fulfil their nutritional requirements (Tadey, 2006). Since rangelands have different proportions of horses, goats, sheep and cows, and that not all livestock produce the same plants damage, we transformed the different livestock densities to animal unit (AU) per hectare, weighting by the history of grazing (i.e. years the rangeland was subjected to grazing), hereafter stocking rate, to make it comparable between rangelands. For this, we considered that 1 cow equals since to 1 AU, 1 horse equals to 1.25 AU, 1 goat equals to 0.17 AU and 1 sheep equals to 0.3 AU, as a modification of the international methodology proposed by Valentine (2001), and as in Souto and Tadey (2018), with stocking rates ranging from 0.06 to 1.63 AU × Years Ha−1 (Table A.1, Appendix A). Previous studies in this area showed that stocking rates evaluated as here are positively and strongly associated to mean browsing percentage (R2 = 0.96; P = 0.0006) and dung density (R2 = 0.66; P = 0.049) while are negatively associated with plant cover (R2 = 0.98; P = 0.04), density (R2 = 0.98; P = 0.02) and richness (R2 = 0.97; P = 0.04); evidencing that for these rangelands, stocking rate is good predictor of plant damage by livestock. In addition, soil nutrient content (carbon, nitrogen and phosphorus) was not correlated with stocking rate (all P > 0.40) (Tadey, 2006).
Studied speciesWe selected Prosopis alpataco var. alpataco (Fabaceae, Mimosoideae) as model species, since it is a shrub highly consumed by livestock and a key species in Monte Desert vegetation (Catalano et al., 2008; Villagra and Roig, 2002). This species is endemic of Argentina and inhabits from 30 to 42°South latitude (Burkart, 1976; Cariaga et al., 2005; Villagra and Roig, 2002). It is characterized for being a spiny shrub that forms circular patches up to 10 m in diameter and 3 m high, its basal branches are always buried, while the secondary branches are arched reflexuous and aerial (Burkart, 1976; Correa, 1984; Villagra and Roig, 2002). P. alpataco is insect-pollinated and has dense and yellowish flower clusters, with a high mean number of flowers per inflorescence (i.e. 131 ± 5 flowers) and presents a high flower mortality between anthesis and fruit set with an average of 28.4 ± 1.3 (% flower × 0.40 m−2) (Cariaga et al., 2005; Chiappa et al., 1997). Therefore, the maximun number of pods per inflorescence usually formed are between 6 to 10 (pers. obs.). Each flower may produce a nutritional legume, with sweet flavor, pale yellow to violet, 7−17 cm long and 0.6–1.2 cm wide and 5 mm thick, with scarce and bitter mesocarp and ellipsoid seeds (Aguero, 2009; Burkart, 1976; Correa, 1984; Villagra and Roig, 2002). Fruit productivity per individual varies between 0 and 4 kg (Vega Riveros, 2009, Vega Riveros et al., 2011), complicating the estimation of total number of flowers and fruits per plant. Seeds present physical dormancy imposed by the hard and waterproof seminal cover (Villagra, 1995). In arid zones of Argentina, Prosopis seeds are dispersed by native and non-native mammals through endozoochory (Campos et al., 2011). This type of dormancy would allow temporal and spatial asynchrony of germination, which represents an advantage in unpredictable dry environments and an adaptation to the endozoic seed dispersal (Vega Riveros et al., 2011). The genus Prosopis comprises 45 species, including shrubs and trees that are important ecological, genetic and economic resources in arid ecosystems, particularly known for producing hard wood and their seeds are an important food source for humans and animals, also being a key element for restoration practices (Bessega et al., 2005; Boeri et al., 2017; Ferreyra et al., 2007; González Galán et al., 2008; Mazzuca and Balzaretti, 2003; Moncada et al., 2019; Pasiecznik et al., 2001; Silva et al., 2000; Villagra et al., 2010; William and Jafri, 2016). Prosopis sp. are key component of the habitat, altering environmental conditions under their canopies, acting as “nurse” species, as they enhance the formation of “fertility islands” through the accumulation of organic matter by fallen brush and enhancing nutrient recycling by their symbiotic interaction with soil microorganism (Aguero, 2009; Fredericksen et al., 2000; Pugnaire et al., 1996; Rossi and Villagra, 2003; Vega Riveros et al., 2011; Villagra et al., 2010). Prosopis is a phylogenetically ancient group and has a worldwide distribution, being Argentina the country with highest number of species (28) with 13 endemic ones (Burkart, 1976) and 8 occurring in Patagonia (Correa, 1984).
Collection and measurementsIn each rangeland, we selected and measured 20–30 individuals, totaling 284 plants. Within each rangeland, individual were sampled randomly, attempting to embrace the largest area possible (∼25 km2 variable depending on the rangeland). Sampling area was located at least 300 m from the road and more than 3 km from the house-farms to avoid additional anthropogenic disturbances. From each individual we collected fresh leaves and fruits to obtain the offspring cohort. We measured plant height and crown diameter (in meters) with a centimeter. We also estimated a browsing index (as a visual estimate of the proportion of the browsed branches per plant). Once mature, we collected an average of 15 fruits per plant, and in the laboratory, we separated seeds from the legume and weighted them in an analytical weighing scale. We considered seeds to be viable when no insect damage was observed and seeds were well formed. With the number of viable seeds we estimate seed set as the number of viable seeds/total seeds per pod. Viable seeds were mechanically scarified with sand paper to break dormancy and promote germination (Vilela and Ravetta, 2001). We sowed three viable seeds per plant in sowing plugs (72 cells) on an organic amendment substrate under greenhouse conditions. Mean greenhouse temperature varied between 18 and 28 °C during the spring-summer period and between 15 and 2 °C along autumn to winter. The seeds were watered weekly to ensure humidity as needed. We monitored seedling emergence every two days during two months and recording along the survey seedling mortality. As a proxy of parental performance, we calculated for each sampled individual the following variables: mean seed set (number viable seeds/total numbers of seeds per parental plant), mean seed weight (in grams) and mean of seedling emergence per parental plant (i.e. number of emerged seeds/ total number of sowed seeds per parental plant), as indirect estimations of fitness through seed quality. We calculated the proportion of viable seeds/pod. Then we estimated the mean proportion of viable seeds per plant, as an estimation of seed set. In this case, each pod corresponds to one flower. In the rangeland with the highest grazing intensity (1.632 AU × Year Ha−1), not all plants produced fruits and, therefore, few legumes could be collected, obtaining no viable seeds and no offspring cohort (Table A.3, Appendix A).
To estimate offspring genetic variation, we collected fresh foliar tissue from 238 seedlings growing under greenhouse conditions. Since the number of seedlings varied among rangelands, we included from 11 to 30 individuals depending on availability, attempting to select them from different parental individuals within each rangeland. All fresh leaf material was preserved in the freezer at −21 °C until we performed genetic analyses.
Isoenzyme methodTo genotype both parental and offspring cohorts we used horizontal isoenzyme electrophoresis technique. Isoenzymes are codominant markers, ideal for population genetic studies of the causes and effects of genetic variation within and between populations. They have a lower level of polymorphism as they have fewer alleles per locus compared with other markers, being superseded by DNA-based approaches, but they are still among the fastest and cheapest marker systems for non-model species, being a cost-effective choice for analyzing many individuals within many populations. In the laboratory we extracted proteins from preserved fresh material using mortars and enzymes following Mitton et al. (1979) methodology. Homogenates were frozen at −80 °C until electrophoresis was performed. Then homogenates were absorbed onto Whitman No. 3 paper wicks, which were run in 12% w/v starch gels. Horizontal electrophoresis was carried out using two buffer systems (morpholine-citrate (MC) and histidine-EDTA buffer), (King and Dancik, 1983; Ranker et al., 1989). In MC system the following enzymes and their putative loci were resolved: Malate dehydrogenase (Mdh-2), Malic enzyme (Me-2, Me-3) and Peroxidase (cathodic: Per-2). Whereas, in histidine-EDTA system the following enzymes and putative loci were resolved: Phosphoglucomutase (Pgm), Phosphoglucosiomerase (Pgi-2, Pgi-3) and Peroxidase (anodic: Per-2). Electrophoresis was carried out at 4 °C until the front indicator, bromophenol blue, migrated approximately 10 cm from the origin to the anode. The cathodic and anodic portions of each gel were sliced horizontally on which the stains suspended in 1% p/v agarose were poured according to Mitton et al. (1979). The bands were stained for specific enzymes, suspended in 1% agar, following standard procedures (Soltis et al., 1983). Band patterns were visualized using a transilluminator. Alleles were numbered sequentially, with the lowest number assigned to the most anodic isoenzyme (Souto and Premoli, 2003).
Data analysisEstimation of genetic parametersTo test neutrality of the isoenzyme loci, we performed the Ewens–Watterson with the POPGENE program version 1.31 (Yeh et al., 1999) and the algorithm given in Manly (1985). To get sufficient precision, the probability was calculated using 1000 simulations. These tests showed that the 8 loci included in this study are neutral (Appendix A, Table A.6). We estimated genetic diversity parameters for parental and offspring populations at rangeland level from isoenzyme data. In the forthcoming analyses we included the following variables: mean number of alleles per locus (A), effective number of alleles per locus (Ae), percentage of polymorphic loci (%P, 95% criterion), observed heterozygosis (Ho) and expected under Hardy-Weinberg equilibrium (He), inbreeding index (Fis) using the Microsoft Office EXCEL plugin GenAlEx v. 6.5 (Peakall and Smouse, 2012, 2006). In addition, we assessed the change in genetic diversity from parental to offspring generation (ΔGV = 1 − parental GV /offspring GV) (Vigouroux et al., 2002), where GV is each genetic variation parameter, where: ΔA is the change in mean number of alleles, ΔAe is the change in mean effective number of alleles and ΔHo, ΔHe are the change in observed and expected heterozygosity, respectively. A relative genetic diversity loss from parental to offspring generation would be represented by a negative value of ΔGV. In addition, we estimated genetic structure using Fisher’ statistics: within populations inbreeding (FIS), among populations genetic divergence (FST) and total genetic structure (FIT), using FSTAT software (Goudet, 1994). In order to detect differences of grazing effect on population genetic structure, we grouped similar stocking rates into three categories as follows: low (L) (0.06 to 0.07 AU × Year Ha−1), intermediate (I) (0.10 to 0.21 AU × Year Ha−1), and high (H) stocking level (0.63 to 0.1.63 AU × Year Ha−1). This grouping allows the genetic structure comparisons within and between stocking level (Table A. 2, Appendix A). Furthermore, we also estimated individual genetic parameter for parental and offspring seedlings within each rangeland from isoenzyme data to be used in a path analysis: individual heterozygosity (IndHet), i.e. number of heterozygous loci divided by total number of loci analyzed per individual, using the Microsoft Office EXCEL plugin GenAlEx v. 6.5 (Peakall and Smouse, 2012, 2006) and individual allelic diversity, i.e. number of alleles of each individual divided by total number of alleles in each population (Peterson et al., 1998; Thoß et al., 2011).
Statistical analysisEffect of grazing on plant fitnessTo investigate grazing effects on parental fitness we used a regression approach using mean plant height, diameter, seed set and seed weight as response variables with stocking rate as explanatory variable. The sampling unit was the individual plant. Models had parental individuals identity nested in rangeland as random factor. Mean plant height and diameter and mean seed weight were analyzed by simple regressions. We transformed plant height and diameter to their logarithm to achieve normality. Seed set and seedling emergence were analyzed with generalized linear mixed models (GLMM). We analyzed seed set with a Beta distribution (used for modelling rates and proportions) (Ferrari et al., 2010) with a logistic link function using the “GlmmADMB” package (Skaug et al., 2018). Whereas for seedling emergence we used a Binomial distribution with logit link function in “lme4” package (Bates et al., 2019, 2014). All statistical analyses were performed using R 3.4.4 (R Development Core Team, 2017).
Grazing effect on genetic parameters in both generationsThe effect of grazing on genetic variation both in parental and offspring cohorts, was analyzed using linear regression models with all the estimated genetic parameters for each rangeland (A, Ae, Ho, He, %P) as dependent variables versus stocking rate, independently. Additionally, we analyzed the relative change of each genetic parameter (ΔA, ΔAe, ΔHo, ΔHe) with stocking rate (explanatory variable), using linear and quadratic regressions. We compared FIT, FST and FIS among stocking levels (low, intermediate and high) and between cohorts with a Kruskall Wallis’s test.
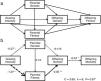
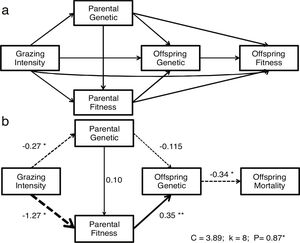
Path analysis: D-separationTo estimate how grazing affect offspring fitness through parental and offspring genetic diversity and parental fitness, we hypothesized a multivariate causal model using path analysis (Fig. 1a). The path analysis was tested with a generalized multilevel “D-separation” test (or “D-sep”) (Shipley, 2009). This method allows estimating causal and non-causal components from total variation through modeling how the variables are linked together by direct and indirect effects leading to a “box and arrow’’ diagram showing how causal effects should flow through the studied system (Shipley, 2009). In this diagram arrows depict direction of causal relationships between variables and the slope of each regression depicts the strength of that relationship (Shipley, 2016). The D-sep method has the advantage that can be used for non-normally distributed data, non-linear functional relationships and data with hierarchical structures (Shipley, 2016, 2009, 2000). Our model tested whether grazing gradient (GG) directly affect offspring fitness (OF) or indirectly through offspring genetic diversity (OG) and/or parental fitness (PF) and parental genetic diversity (PG). In turn PG may directly affect PF and OF and OG. Additionally, PF and OG may have an effect on OF (Fig. 1a). In this model, we used stocking rate as GG proxy. To summarize “genetic diversity” and “parental fitness” in a single integral variable, we performed three Principal Component Analysis (PCA): one using the IndHet and individual allelic diversity for parental genetic diversity, another with the same variables for offspring, and a third PCA using viable seeds proportion, seed weight and seedling emergence as proxies of parental fitness. All these analyses were performed with variables estimated at individual level within each rangeland. In further analyses, we used first PCAs’ axis scores explaining more than 60% of the variance. As a proxy of OF, we used seedling mortality. Individual seedling mortality was analyzed with a Bernoulli distribution recommended for binary variable where 0 represents seedling survival and 1 is seedling mortality (Zuur et al., 2009). Models with seedling mortality had individual nested in rangeland as a random factor. The other analyses were performed using linear regressions (Table A.4, Appendix A). To test the path model we first explored the existence of independence among variables (claims, Table A.5, Appendix A) sensu the hypothesized model of Fig. 1a. Then to test whether the model fits the data, we estimated parameter C following the equation:
where k is number of independent claims, Pi is null probability for each of k. We then compared resulting C value to a χ2 distribution with 2k degrees of freedom (Shipley, 2016, 2009, Shipley, 2000). We rejected the causal model if the C value is unlikely to have occurred by chance (i.e., P < 0.05) (Shipley, 2009). The association between each pair of variables depends on both direct and indirect relationships (Legendre and Legendre, 1998). To obtain total indirect effect of stocking rate on offspring fitness, we multiplied estimated coefficients for each pair of variables involved in each possible indirect pathway. Then, we summed all indirect and direct pathways effects to estimate total covariation (r) (Legendre and Legendre, 1998; Shipley, 2000). There were three indirect pathways from GG to OF. 1) From grazing gradient to parental genotype, from parental genotype to offspring genotype and from offspring genotype to offspring fitness (GG-PG-OG-OF). 2) From grazing intensity to parental genotype, from parental genotype to parental fitness, from parental fitness to offspring genotype and from offspring genotype to offspring fitness (GG-PG-PF-OG-OF). 3) From grazing gradient to parental fitness, from parental fitness to offspring genotype and from this one to offspring fitness (GG-PF-OG-OF) (Fig. 1a).Causal model describing grazing effects on parental and offspring fitness and genetic diversity. a) Hypothetical model. b) Significant model: continuous arrows indicate positive effects; dashed arrows indicate negative effects and arrow thickness indicate the strength of the relationship. Regression coefficients are near arrows with their significance level as follows: ·P < 0.10, *P < 0.05, **P < 0.005. The suitability of the models was assessed on the basis of χ2 and the associated P values (a higher P value means a better fit of data with the model).
Based on D-sep results, we analyzed the strongest path. To explore how parental fitness affected offspring genetic population parameters, we subjected these data to separate linear and polynomial regressions (quadratic term; taking into account the offspring genetics quadratic response to grazing), analyzing effective number of alleles (Ae), expected heterozygosity (He) and inbreeding (FIS) as dependent variables, and parental proportion of viable seeds, seed weight and seedling emergence as independent variables (fixed effects).
ResultsEffect of grazing on plant fitnessWe found that grazing intensity linearly reduced the seed set (β = −2.5, P < 0.0001) and seedling emergence (β = −1.64, P < 0.0001). The browsing index tended to increase with increasing grazing intensity (β = 0.59, P = 0.11), while plant height, diameter and mean seed weight did not show significant responses to grazing (all P > 0.14).
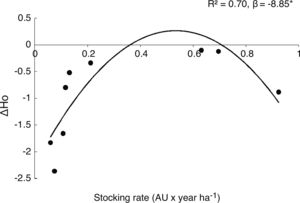
Grazing effect on genetic parameters in both generationsWe observed that as grazing intensity increased, genetic diversity of the parental cohort linearly decreased in mean number of alleles (A) (β = 0.42, P = 0.016), observed heterozygosis (Ho) (β = −0.10, P = 0.017), expected heterozygosis (He) (β = −0.12, P = 0.005) and percentage of polymorphic loci (%P) (β = −0.25, P = 0.014). Regarding offspring cohort, we found that grazing did not linearly affect population genetic parameters (all P > 0.5). However, Ae and Ho showed a negative quadratic association with stocking rate (β = −2.42, P = 0.05; β = −0.78, P = 0.017, respectively). In addition, when analyzing the differences in population genetic parameters between cohorts (parental-offspring), we found that stoking rate showed a significant and negative quadratic association with ΔHo (β = −8.85, R2 = 0.70, P = 0.030) and marginally with ΔA (β = −1.18, P = 0.08), ΔHe (β = −6.88, P = 0.10) and ΔAe (β = −2.48, P = 0.11); (Fig. 2 and Fig A.2, Appendix A).
Relative genetic loss between cohorts along a grazing gradient. Dots represent changes in observed mean heterozygosis (ΔHo= 1- parental Ho /offspring Ho) between parental and offspring population for each rangeland. Negative values of ΔHo show the loss of Ho from parental to offspring cohort. *denotes P < 0.05.
We found that total genetic structure (FIT) of parental cohort in rangelands with low grazing intensity was not different from zero and was mainly explained by divergence between populations (FST > FIS, Table 1). As stocking level increased, FIT in parental populations turned significantly different from zero and more influenced by within population inbreeding (FIS), although at high stocking levels the influence of FIS was similar to between populations divergence (FST) (Table 1). None of these parameters show differences among stocking levels (FST χ2 = 1.14, P = 0.56; FIS χ2 = 0.29, P = 0.87). Offspring cohort, instead, showed high levels of total genetic structure (FIT) in rangelands with low grazing intensity, which is almost completely explained by within population inbreeding (FIS > FST, Table 1). Whereas as stocking level increased, total genetic structure decreased (FIT) and is similarly explained by both, within population inbreeding and among population divergence (FIS ∼ FST, Table 1, Fig A.3, Appendix A).
Mean of F-statistics (FIT, FST, FIS) and their confidence intervals for each stocking level (low, intermediate, high) and cohort (parental and offspring). ns = not significantly different from zero. * = significantly different from zero.
| Stocking level | Cohort | FIT | FST | FIS |
|---|---|---|---|---|
| Low | Parental | 0.021 | 0.041 | −0.021 |
| (-0.08−0.114) ns | (0.009−0.073)* | (-0.124−0.074) ns | ||
| Offspring | 0.406 | 0.09 | 0.347 | |
| (0.293−0.511)* | (0.042−0.119)* | (0.225−0.466)* | ||
| Intermediate | Parental | 0.246 | 0.063 | 0.192 |
| (0.116−0.351)* | (0.03−0.109)* | (0.058−0.306)* | ||
| Offspring | 0.210 | 0.108 | 0.116 | |
| (0.069−0.382)* | (0.015−0.21)* | (-0.06−0.323)ns | ||
| High | Parental | 0.249 | 0.114 | 0.153 |
| (0.162−0.347)* | (0.069−0.164)* | (0.055−0.263)* | ||
| Offspring | 0.238 | 0.081 | 0.114 | |
| (0.132−0.380)* | (0.007−0.148)* | (0.011−0.364)* |
First we tested a hypothetical complete model (Fig. 1a) including all possible relationships among studied variables obtaining a non-significant model. Then, we tested a reduced model that included only those variables with significant effects, obtaining a fitted model that did not reject the null hypothesis (P = 0.87, Fig. 1b). In this model, the grazing gradient reduced parental fitness (PF) (r = −1.27) and genetic diversity (r = −0.27), and ultimately reducing seedling mortality through offspring genetic diversity(r = −0.34). The strongest indirect pathway (r = 0.15) was from grazing gradient, through parental fitness (PF), offspring genotype (OG) to offspring fitness (OF) (GG-PF-OG-OF). This pathway showed a positive effect of grazing gradient on seedling mortality, while the rest showed lower strength (GG-PG-OG-OF, r = −0.01 and GG-PG-PF-OG-OF, r = 0.003, respectively).
Relationships between parental plant fitness and offspring genetic parametersWe found that parental seed set was marginal and quadratically associated with effective number of alleles (Ae, β = −0.47, P = 0.059) and showed a quadratic tendency with expected heterozygosity (He, β = −0.15, P = 0.13) of offspring cohort. Also, that seedling emergence was marginal and quadratically associated with offspring effective number of alleles (Ae, β = −11.34, P = 0.054) and significant and quadratically associated with offspring expected heterozygosity (He, β = −4.98, P = 0.019). Meanwhile, seedling emergence was marginal and linearly associated with offspring inbreeding (FIS, β = −0.61, P = 0.092). Seed weight did not show association with other offspring genetic parameters neither linearly nor quadratic (Ae, He, Fis, all P > 0.32) (Table 2).
Relationships between parental fitness and offspring genetic parameters. Analyses of the effects of parental fitness on offspring genetic diversity. Genetic parameters evaluated were: Ae = effective alleles number, He = expected heterozygosity, FIS = inbreeding index.
| Explanatory variable | Response variable | Regression type | β | P | df |
|---|---|---|---|---|---|
| Seed set | Ae | Quadratic | −0.467 | 0.059 | 7 |
| He | Quadratic | −0.150 | 0.131 | 7 | |
| FIS | Simple | −0.103 | 0.733 | 7 | |
| Seed weight | Ae | Quadratic | −0.092 | 0.346 | 7 |
| He | Quadratic | −0.043 | 0.241 | 7 | |
| FIS | Quadratic | −0.008 | 0.903 | 7 | |
| Seedling emergence | Ae | Quadratic | −11.34 | 0.054 | 7 |
| He | Quadratic | −4.978 | 0.019 | 7 | |
| FIS | Simple | −0.611 | 0.092 | 7 |
Long lasting disturbances on natural populations may cause genetic diversity erosion through generations. We tested whether exotic large herbivores produce transgenerational negative effects on phenotypic and genetic traits of desert vegetation. As hypothesized, we found that grazing mostly affected parental cohort fitness and its genetic diversity and structure, consequently affecting the offspring. Genetic diversity was lost from parental to offspring cohort along the grazing gradient, although the effect was relatively lower at intermediate levels of grazing. As expected, we observed inbreeding effects on all offspring populations, while parental populations showed inbreeding on moderate and highly grazed rangelands. In addition, there was a negative impact of grazing on offspring survival. Finally, we summarized and discussed the causal relationships among grazing intensity, fitness and genetic diversity parameters between cohorts.
We observed that large herbivores fed on P.. alpataco plants and this occurred in all the studied rangelands, with a mean browsing index for all parental populations of 67 ± 30 % (Fig. 3b). This contradicts previous results in the same region that found that browsing index to other species increased linearly with stocking rate (Tadey, 2006). However, this reflects the species high palatability that even in slightly grazed rangelands, with low encounter probability; there were several highly consumed individuals (>40% browsed in average, Table A.3, Appendix A). We found a significant decrease in plant performance in terms of seed set and seedling emergence along the grazing gradient. These results could be explained by the plant damage effects on reproduction, such as the reduction in resources available to invest in reproduction or by direct consumption of flowers, which in turn could affect pollination services, and/or fruits. Inbreeding may also cause an increase in aborted seeds decreasing seed set (Castilla et al., 2019). Aguirrebengoa et al. (2018) found that ungulates significantly reduced seedling emergence rate, attributing it to lower viability by a reduction in seeds carbon content, which could even explain the higher seedlings mortality. On the other hand, large herbivores did not affect plant height and crown diameter; this apparent discrepancy with the high damage observed could be explained by several facts. First, this spiny shrub sprouts radially generating large crowns up to 8 m in diameter, which generally deters grazing. However, in highly grazed rangelands resources for large herbivores become scarce and this plant species is highly palatable. In this context, herbivores can manage to destroy a crown and open a trail inside of it, foraging the plant center and giving it a ring-shape, without varying its total diameter (Fig. 3a–b). Large herbivores usually consume P. alpataco’s small young leaves, flowers and fruits, and less frequently, stems and spiny branches. We suggest the addition of a measurement of green biomass to estimate grazing damage in future studies. Moreover, we observed no variation in seed weight along grazing gradient, suggesting that seed weight is maintained, probably at the expense of reduced seed quantity, through maternal effects mechanisms (Agrawal, 2001). Nevertheless, our results suggest that grazing is affecting, at least partially, reproduction success through plant damage of a xerophytic shrub that depends mainly on sexual reproduction to persist over time.
Consequences of long lasting disturbances on plant abundance and reproduction can affect ecosystem dynamics in different ways that, through time, end up impacting on their genetic diversity and structure (Davies et al., 2016). As mentioned above, genetic diversity is essential for species persistence over time. We observed different negative effects of large herbivores on genetic diversity between cohorts. In parental cohort, we found that grazing reduced genetic diversity, which suggests that large herbivores are eliminating standing individuals (i.e. genotypes) through consumption or trampling. P. alpataco is a long-lived, self-incompatible species (Harris, 2003), and probably sampled individuals existed before the introduction of exotic livestock in the region. Therefore, it is also probably that livestock had been selecting (or rejecting) some genotypes, leading to within population gene pool homogenization. Accordingly, we found increasing inbreeding within and divergence among parental populations with grazing intensity. Moreover, genetic drift may lead to further reductions in genetic diversity (Frankham, 1995). Consequently, a reduced population size and/or cross-reproduction could restrict gene flow, depaupereting the gene pool across cohorts. In this way, few individuals and/or fewer breeding individuals contribute to seed production to future generations and, therefore, part of the population’s genetic diversity is lost (Souto and Tadey, 2018). These detrimental effects of grazing on genetic variation may jeopardize population’s ability to persist in stressful or changing environments across generations (Frankham et al., 2002).
Grazing effects on the offspring cohort were more complex. Genetic diversity of parental to offspring populations is expected to vary due to sexual reproduction; greater outcrossing in parental population would lead to greater genetic diversity in the offspring. However, we observed that as large herbivores increased in abundance, offspring genetic diversity showed a negative quadratic response. Even when genetic diversity was lower in offspring than in parental generation in all studied rangelands (i. e. negative ΔHo values), this loss was lower at intermediate grazing intensities (Fig. 2). These results may allude to the “intermediate-disturbance hypothesis” (Connell, 1978; Wilkinson, 1999), suggesting that in offspring cohort, an intermediate level of disturbance would affect genetic diversity loss less than extreme grazing intensities. This might be explained by pre-zygotic mechanisms, mainly during pollination. A recent study reported that intermediate levels of grazing did not impair bee communities (Shapira et al., 2019). Similarly, in the same rangelands included in this study, pollinator richness and abundance were higher at intermediate than at extreme grazing intensities (Tadey, 2015). This might possibly be the result of a reduction in the abundance of dominant species and the increase of rare ones by reduced competition; this in turn would improve pollination services, favoring cross-pollination at intermediate grazing intensities (Castilla et al., 2019; Lázaro et al., 2016; Lehtilä and Strauss, 1997; Tadey, 2015; Winfree et al., 2007). In accordance with this, we observed lower levels of inbreeding by kinship in the offspring at intermediate grazing intensities. So higher pollen dispersal might be preventing inbreeding depression (Gandon and Michalakis, 1999; Gandon, 1999; Hamilton and May, 1977; Taylor, 1988). To some extent, intermediate grazing could also benefit P. alpataco establishment through a genetic structure determinant, such as seed dispersal (Bessega et al., 2017), by seeded droppings and subsequent trampling and burial. Additionally, since P. alpataco has high germination rates once it escapes herbivory it could easily establish in bare soil where there is less competition (Campos et al., 2011; Paez and Marco, 2000). Nevertheless, there is little information on the role of livestock in the dispersion process of P. alpataco (Vega Riveros et al., 2011).
At the extremes of grazing intensity reproductive success may be limited by the level of kinship among intraspecific neighbors, as generally nearby individuals may exhibit higher relatedness levels (Castilla et al., 2019; Loveless and Hamrick, 1984), and by pollinator behavior that could be affecting plant-pollinator interactions, such as distances of pollen dispersal at which the genetic neighborhoods develops (Breed et al., 2015; Kolb, 2008; Lloyd et al., 2018; Tadey, 2015). In the case of low grazing intensities, where there are patches with more plants density and all species bloom almost simultaneously, there is a greater competition between pollinator species and between plants for pollinators (Tadey, 2006). This could be reducing pollen quantity (pollen limitation) and quality through inbred pollen dispersal due to territorial pollinator foraging behaviors (i.e. foraging only on a reduced group of individuals/patches) (Castilla et al., 2019; Tadey, 2008, 2015). This is supported by the higher inbreeding between cohorts observed in low grazing intensity rangelands. Meanwhile, highly grazed rangelands have lower plant density, cover and species richness (Tadey, 2006), leading to more isolated flowering plants and patches increasing pollinator foraging costs (Kolb, 2008; Tadey, 2020, 2007). Consequently, grazing would be indirectly enhancing self-pollination by limiting pollinator movement among patches because pollinators tend to visit more flowers per plant to reduce foraging costs (Harder, 2014; Jennemten, 1988; Kolb, 2008; Sih and Baltus, 1987; Tadey, 2015). In turn, self-pollinated endogamic pollen may result in non-viable seeds, particularly in those plant species with a self-incompatible reproduction system (Harris, 2003), as observed in our results. Therefore, large herbivores could be affecting pollen dispersal and hence gene flow within plant populations differentially along the grazing gradient, whereas at intermediate levels of grazing better pollination services might decrease inbreeding through generations.
In general, genetic diversity losses are associated with a reduction in fitness due to inbreeding depression, both at the individual and population level, which further reduces the population size (Bouzat, 2010). We observed negative grazing effects on parental cohort, both on phenotypic and genotypic variation, with consequent transgenerational effects on their offspring. We found grazing effects on seedling mortality exclusively through indirect mechanisms. Aguirrebengoa et al. (2018) found some similar results, showing direct negative effects of ungulates on offspring cohort, increasing their mortality rate, ascribing it to a reduction in seed quality by parental plant damage. According to our model, grazing negatively affected parental genetic diversity and fitness, which in turn affected offspring genetic diversity, increasing seedling mortality. Among the three proposed indirect pathways that increase mortality rate (Fig. 1b), the pathway including parental fitness (i.e. PCA scores summarizing three variables, viable seeds, seed weight and seedling emergence) through offspring genetic diversity was the strongest; and as above described, we observed a strong decrease in plant performance along the grazing gradient. Grazing may restrict pollination with consequences on plant species demographic structure and/or spatial and temporal distribution of sexually compatible plants, limiting gene flow by decreasing the likelihood of pollen deposition from unrelated plants (Tadey, 2008) and affecting offspring mortality by inbreeding depression, as we observed here. Usually, higher genetic diversity is associated with an increase in plant fitness (Bouzat, 2010). We observed, in our model, that parental genetic marginally increased their fitness (PCA scores), but greater offspring genetic diversity had negative correlations with seedling mortality. Additionally, we observed a marginal and negative association between parental and offspring genetic diversity. This may result from the high variability observed on genetic diversity (PCA scores) along the grazing gradient (Fig. A.4, Appendix A), different type of responses from each generation to grazing intensity (linear vs. quadratic) or similarities in genetic diversity between both cohorts. Suggesting that one generation might not have enough time to see significant changes in genetic diversity or that a larger sample size is needed. However, our results also showed significant losses of genetic diversity between cohorts with long-term effects. The path analysis evidenced that the main mechanism by which grazing affected offspring genetic diversity and survival was through parental fitness. The negative and quadratic associations among seed set and seedling emergence of parental cohort on the effective number of alleles and expected heterozygosity of offspring cohort showed that populations with intermediate production of viable seeds and emergence rates produce offspring with higher genetic diversity (Ae and He). This could be limiting offspring gene pool, whereas at intermediate grazing intensities, with intermediate viable seeds production and seedlings emergence, there is greater genetic diversity in offspring, possibly due to greater richness and abundance of pollinators (i.e. better pollination services). It would be useful to confirm these results with another type of more sensitive and more polymorphic molecular markers than isoenzymes, such as microsatellites. Our results also highlight the need of measuring reproduction success both in quantity and quality, in terms of cross-breeding with non-inbred pollen, seedling vigor and survival as we found that evidenced with the greatest seedlings emergence occurred in populations with less inbreeding (FIS). Since parental fitness was highly associated to parental genetic diversity, we may assume that parental genetic diversity is also important for offspring persistence over time. In this sense, indirect grazing impacts on offspring fitness are expected to be much greater in arid field conditions, compromising natural establishment and exacerbating genetic diversity losses over generations (Pol et al., 2014; Sternberg et al., 2003; Tadey and Souto, 2016; Tadey, 2007; Trlica and Rittenhouse, 1993). In consequence, a decrease in species fitness can decrease population growth rate and potentially decrease their sizes with greater losses of genetic diversity over time, consequently affecting their adaptive potential to environmental and/or anthropogenic changes (Hendry et al., 2018; Lande, 1994; Lynch et al., 1995).
Therefore, the maintenance of genetic diversity needs to be encouraged for the long-term sustainability of unmanaged dryland rangelands (Souto and Tadey, 2018), where extreme environmental conditions make them more susceptible to disturbance than other more humid ecosystems. Some of the measures that could mitigate population reduction are the inclusion of sustainable management practices such as livestock rotation, transhumance practices and the application of ecological restoration. In the case of the latter, increasing biological and genetic diversity should also consider the type of species with different growth rates, such as early or pioneer species, which may be less susceptible to grazing and have a faster recovery rate than late species, have high reproductive success, provide high availability of resources for pollinators, consider their phenology and seed dispersal.
ConclusionsGenetic diversity enables species to persist over time by overcoming disturbance effects. This study shows that grazing by large exotic herbivores can change gene flow in different pathways depending on the intensity, with consequences on genetic diversity and structure. We observed that offspring populations showed significant inbreeding effects. We found that genetic diversity loss between cohorts involved complex, non-linear and indirect mechanisms. Generally, genetic diversity losses were associated with a reduction in parental fitness, which in turn affected offspring genetic diversity and increased seedlings mortality. Hence, studies along gradients of disturbance and understanding relationships among variables in a causal framework are important to detect these processes. Our results highlight the relevance of estimating intra and inter-generational direct and indirect of long-term effects of grazing on plant population dynamics. Finally, the introduction of large exotic herbivores in unmanaged rangelands are seriously affecting dryland vegetation structure, with substantial consequences on ecological processes and ecosystem services. Livestock grazing may be, therefore, jeopardizing desert species evolutionary potential, compromising natural revegetation and aggravating genetic diversity losses along generations that might be deepened under forecasted climate change.
Declaration of interestsThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors thank Hugo Saiz for helpful comments on an early version of the manuscript, and two anonymous reviewers and Editor for their helpful recommendations that have significantly improve it. This study was supported by projects PICT 2014-3478, Argentine Science and Technology Secretary and PIP-2015-2017, CONICET.