There is an urgent need for rapid, standardised, accurate and accessible monitoring techniques to better detect and quantify change given the increasing threat of degradation and biodiversity loss in freshwater ecosystems. Community-based monitoring projects have been proven successful for the collection of meaningful biological data from a range of target species and ecosystems. The STREAM (Sequencing the Rivers for Environmental Assessment and Monitoring) project combines community-based monitoring with a DNA metabarcoding approach to assess aquatic ecosystem health by determining biodiversity of benthic macroinvertebrate species across Canadian watersheds. STREAM consists of outreach and recruitment, training and dissemination of results obtained from sequence data, allowing rapid generation of watershed biodiversity reports (e.g. in 2 months). We emphasise the benefits of partnering with community groups in these DNA biomonitoring efforts, highlighting the value of environmental stewardship and eliminating bottlenecks for scientific data collection. We believe the approach taken in STREAM is not only applicable to Canada, but functions as an ideal model for freshwater monitoring on a global scale.
Climate change and additional anthropogenic stressors, including habitat loss and pollution, are currently driving regional and global shifts and deterioration in species distribution, composition and abundance (Butchart et al., 2010; Brown et al., 2016; Jackson et al., 2016; Warren et al., 2018). The ability to detect and understand these global changes is of vital importance for preventing further biodiversity loss (Butchart et al., 2010; Anderson, 2018; Kissling et al., 2018). In a rapidly changing world, faster and more diagnostic biomonitoring techniques are urgently required for accurate assessment of modifications of ecological state (i.e. biodiversity loss), in global ecosystems (Butchart et al., 2010; Jackson et al., 2016).
Freshwater ecosystems provide vital ecosystem services to humans and are critical for the wellbeing of many species (Mittermeier et al., 2010; Darwall et al., 2018). With human populations expected to grow to 9 billion by 2050 (Gleick and Palaniappan, 2010), pressures on freshwater ecosystems are intensifying. Freshwater scarcity and security are only two of many major issues behind the global freshwater biodiversity crisis (Vörösmarty et al., 2000; Jury and Vaux, 2005; Srinivasan et al., 2012; Darwall et al., 2018), and in the Anthropocene, demand for freshwater will only increase, thus accelerating degradation and overall negative impacts on freshwater (Waters et al., 2016; Darwall et al., 2018) unless ecosystems are managed wisely.
Monitoring freshwater ecosystems in the face of local anthropogenic effects and global climate change has proved to be challenging (Geist, 2015; Hermoso et al., 2016; Jackson et al., 2016). Despite only covering 1% of the Earth's surface area (Mittermeier et al., 2010), freshwater habitats are widespread (Moss, 1994), often inaccessible either physically or due to political barriers (Salwasser, 1990; Conrad and Hilchey, 2011). Overcoming these obstacles is crucial to enable representative assessment of freshwater health at appropriate spatial scales (Jackson et al., 2016). Public participation in biological data collection can play a crucial role in addressing physical and social barriers to ecological data collection and consequently, community water-based monitoring (CWBM) has grown significantly in the last decade (Conrad and Hilchey, 2011; Gura, 2013; Pocock et al., 2015; Weston and Conrad, 2015). This activity, widely termed as a ‘citizen science’ or ‘community-based monitoring’, has been used globally as an observation and data collection approach for both long-term and focussed short-term projects and is increasingly recognised as a vital component of environmental protection and management (Greenwood, 2007; Dickinson et al., 2012; Biggs et al., 2015; Environmental Protection Belongs to the Public: A Vision for Citizen Science at EPA, 2017; Heigl et al., 2019). In Australia, the Tasmanian State Government has conducted broadscale monitoring of river condition, using the Australian River Assessment System (AusRivAS) protocols, which focuses on macroinvertebrate communities and habitat quality (Smith et al., 1999). In Canada, the large-scale Canadian Aquatic Biomonitoring Network (CABIN), was implemented to provide a consistent approach to biological assessment and to support the aquatic biomonitoring needs of a diverse array of network partners. CABIN has been working with community groups for the last 20 years to collect and analyse data on freshwater health, by providing standardized sampling protocols, training, shared database and mapping tool, and common data interpretation methods based on a Reference Condition Approach (RCA) for assessing aquatic ecosystem condition (Environment and Climate Change Canada, 2012; Jones et al., 2011).
Quite often with community-based biomonitoring research, community groups with an interest in the biological questions being asked are approached as potential collaborators for environmental monitoring, including direct sample and/or data collection (Dickinson et al., 2012). This approach can facilitate wider involvement in regional or national monitoring activities, while increasing the volume of data collected (Dickinson et al., 2012). The involvement of community residents in generating biological information goes beyond research (Phillips et al., 2019); by forming such collaborations, community groups are empowered as volunteer field technicians and enhance their scientific literacy (Bela et al., 2016). Community-based projects result in improved science–society–policy interactions and drive a more bottom-up democratic approach to knowledge-generation based on evidence and informed decision making (Bela et al., 2016; Hsu et al., 2017; Hecker et al., 2018). Successful community-based projects are often executed within a framework which includes training of volunteers, optimised data processing methodology and accessible dissemination of project results (Bonney et al., 2009; Silvertown, 2009; Conrad and Hilchey, 2011; Dickinson et al., 2012; Newman et al., 2012). Frameworks involving these three elements are crucial for consistency and reproducibility of nationally focused projects for trans-national or global scales of biological monitoring.
Supporting program operation through standardised protocols for environmental sample collection linked to training support is a key element in the development of DNA-based community-based monitoring projects (Biggs et al., 2015; Deiner et al., 2017; Environment and Climate Change Canada, 2018). Design and optimisation of commercial sampling kits and bioinformatic pipelines have facilitated the involvement of community groups in biodiversity research, reduced monitoring costs and sped up the data generation process (Biggs et al., 2015; Deiner et al., 2017; Leese et al., 2018; Compson et al., 2020). Such automated DNA-based approaches for biomonitoring empowers communities and draws attention to the issue of biodiversity declines from local to nationwide scales.
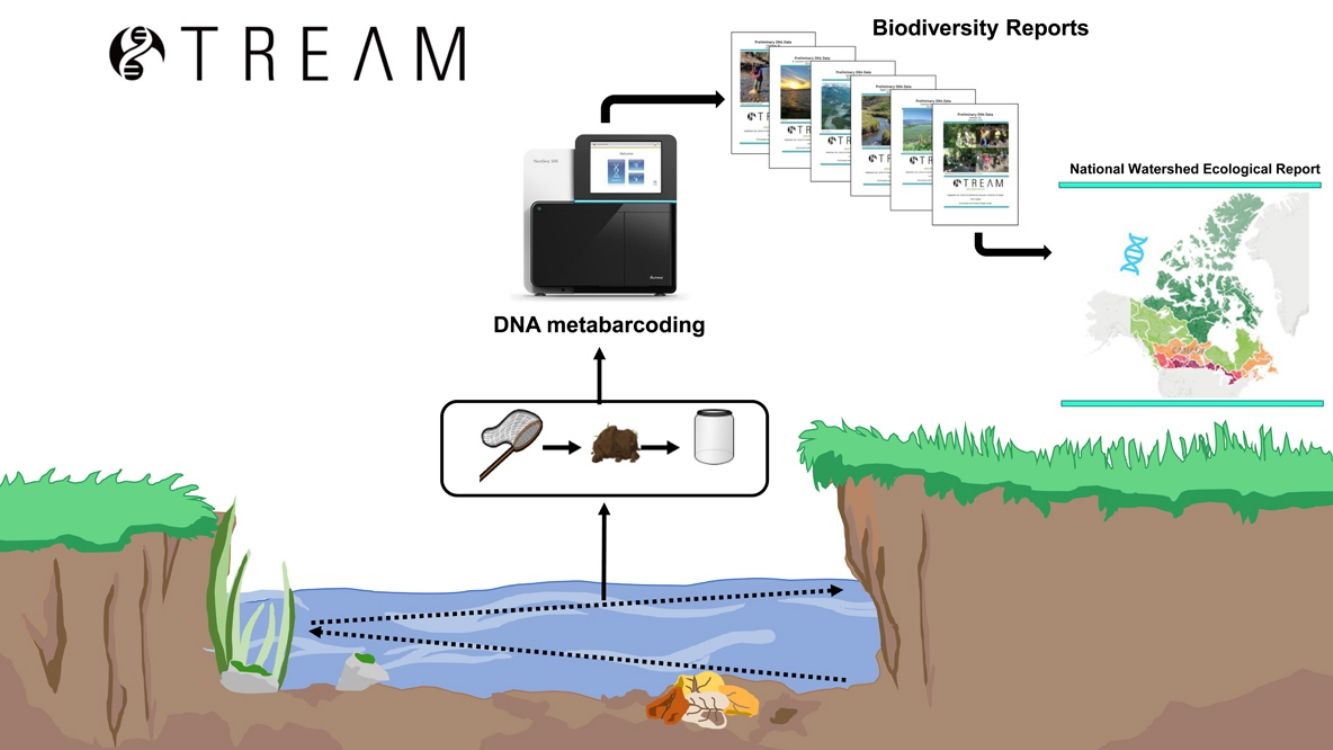
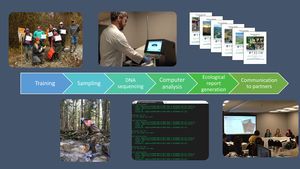
STREAM (Sequencing the Rivers for Environmental Assessment and Monitoring), is a biomonitoring project which involves the combination of community-based monitoring and DNA metabarcoding technologies to assess aquatic ecosystem health using benthic macroinvertebrate communities in watersheds across Canada (www.stream-DNA.com). DNA metabarcoding comprises the mass-amplification of DNA from a range of target taxa (i.e. macroinvertebrates) from an environmental sample (i.e. benthos). Built on over a decade of research and development in applying DNA metabarcoding, coined Biomonitoring 2.0, for rivers and wetlands benthic analysis (e.g. (Hajibabaei et al., 2011; Baird and Hajibabaei, 2012), STREAM is a national multi-stakeholder collaboration between the Centre for Biodiversity Genomics at University of Guelph, World Wildlife Fund (WWF) Canada, Living Lakes Canada (LLC), and Environment and Climate Change Canada (ECCC). This unique merging of academic and non-academic partners facilitates community and bioinformatic processing, with a government platform, CABIN for networking and outreach (Fig. 1). STREAM was established with the main premise of collecting 1500 samples across 15 watersheds over a 3-year period. Watersheds to be targeted include some which are currently being monitored through the CABIN network, together with other data-deficient watersheds. STREAM, via Living Lakes Canada, aims to train 40 new community-based monitoring volunteers, of which this number has already been exceeded, with 100 new trained volunteers recorded in February 2020. In terms of data processing and reporting. The overarching research question aims to address whether we can better understand freshwater ecosystem health across Canada, through applying diversity, functional and richness metrics of macroinvertebrate DNA samples collected in conjunction with community-based biomonitoring groups. STREAM aims to fast-track identification and classification of macroinvertebrates from up to 6-12 months (referring to sample collection, identification, quality assurance/quality control and report generation; (Gardner et al., 2008; CABIN, 2014; Hajibabaei et al., 2016)) to 1-2 months. The aim of this is to increase the taxonomic and spatial coverage of sampling locations, creating the potential for more comprehensive interpretation and diagnosis of environmental condition.
Here we discuss the framework of the STREAM project and explore how we can apply this community-based project approach beyond Canada for global freshwater monitoring.
Partnerships with community groupsUnderstanding motivations of communities to participate in community-based monitoring projects is important for ensuring initial uptake and data collection throughout the duration of the project (Jordan et al., 2012). It is expected that benefits for the communities (Jordan et al., 2012) and project incentives (Dickinson et al., 2012) are fundamental drivers of public engagement (Rotman et al., 2014). Duration of project and frequency of data collection required can also be determining factors for uptake and sustainability of engagement (Silvertown, 2009; Rotman et al., 2014). Often, the greatest influence for involvement with biological monitoring projects is the research question being addressed with the data collection (Bonney et al., 2009; Dickinson et al., 2012; Tweddle et al., 2012; Rotman et al., 2014). Data collection volume often forms the first ‘bottleneck’, regarding the impact of the research findings (Silvertown, 2009). Therefore, it is imperative to form partnerships with community groups that have a vested interest in the implications of the research and have the capabilities to collect data long-term.
Project interest is a fundamental component to building a successful community-based programme. STREAM acknowledges this and therefore aims to partner with groups of individuals which either have prior experience or training with freshwater monitoring and/or have a vested interest in addressing their own ecological questions within the scope of STREAM. In Canada, collaborating with First Nations, Inuit and Métis (referred to as FNIM hereafter) as stakeholder groups for study design and sample collection is important on two levels; firstly, this helps to build community technical capacity and secondly, this facilitates the co-creation of data and provides the opportunity to learn from Indigenous Knowledge (Pocock et al., 2019). This approach ultimately facilitates two-eyed seeing (Bartlett et al., 2012), which is essential in the context of environmental protection and management (Absolon, 2011). Working in conjunction with NGOs such as WWF-Canada and Living Lakes Canada, STREAM facilitates the networking of FNIM groups with researchers to achieve a common goal of a better understanding of freshwater biodiversity. As described by Nerbonne and Nelson (2008), being part of a network of engaged biomonitoring communities encourages data use, and STREAM partnerships with Environment & Climate Change Canada's CABIN network and Living Lakes Canada, can facilitate receptor uptake and data application within engaged communities.
STREAM considers community groups involved in this project as partners, who have a vested interest in identifying their local biodiversity for the purpose of preserving/sustaining it as a valuable local resource. As well as the benefits of data collection for assessing freshwater health, there are numerous benefits to the communities themselves, including: hard data, assistance with data interpretation, and the ability to leverage this information to advocate for protections/management interventions to sustain their local freshwater resources. It is important to highlight that STREAM gives back to local communities, in the form of empowering local communities, facilitating the learning of new skills and providing an open channel of communication between community groups and researchers.
Education, training and sample collectionAn essential component of this biomonitoring approach is a robust training element, which covers all aspects of sample collection that is realistic and achievable by non-specialists and communities alike (Gardiner et al., 2012; Tulloch et al., 2013; Bonney et al., 2014). Sample protocol complexity is known to be a key determining factor as to whether community-based biomonitoring projects are successful (Bonney et al., 2009; Parsons et al., 2011; Crall et al., 2013). Often, the higher the effort input required for biological sampling, the greater the likelihood that communities will cease engagement with biomonitoring projects (Bonney et al., 2009; Parsons et al., 2011). However, the use of visual aid such as training videos and online materials can aid the training process (Newman et al., 2012), even for multi-step protocols such as in Biomonitoring 2.0.
STREAM employs the existing CABIN framework, developed and led by Environment and Climate Change Canada, to train community groups in the Biomonitoring 2.0 approach. Environment da and Living Lakes Canada undertake extensive outreach activities to promote training events, via websites, newsletters and through directly communicating with community groups and stakeholders. The existing CABIN network functions effectively as a method of information communication and wider dissemination. Elements of CABIN online training include foundation modules, such as ‘Introduction to Biomonitoring’, which includes resources on bioindicators and aquatic ecology, modules on site selection and sample collection and focused module components on analytical methods and how reports are used in site assessment. The practical elements of this training are delivered by Living Lakes Canada staff and covers all aspects of sample collection, decontamination and shipping, with end users receiving a certification on completion of field training and passing of module quizzes.
CABIN training and sample collection protocols have been implemented within community groups with great success for over 20 years (Buss et al., 2014; Strachan and Reynoldson, 2014; Gibson et al., 2015), and only a slight modification to sample processing post-collection is required for DNA metabarcoding. For STREAM, river/stream samples are collected via the standardized CABIN Wadeable Streams Field Protocol (Environment and Climate Change Canada, 2012) and wetland samples through the CABIN Wetland Macroinvertebrate protocol (Environment and Climate Change Canada, 2018). The addition of ethanol or antifreeze preservative after sample collection is the only element which differs from the original protocol. This approach of working in conjunction with non-academic organisations and following a standardised training programme, efficiently utilises expertise of training programme coordinators and facilitates high-quality, reliable and comparable data collection.
Facilitating remote and multi-taxa samplingTo achieve the desired broad spatial coverage of STREAM samples and to facilitate the generation of biomonitoring from multiple taxa of interest, we have previously developed and optimised certain elements of sample collection and storage. Across Canada, many of the data-deficient watersheds for freshwater health are located within remote regions (WWF-Canada, 2017). High purity (e.g. molecular biology grade) ethanol is commonly used to preserve benthos samples, however as this chemical is classified as hazardous, challenges arise regarding storage and transit of benthos samples preserved in ethanol (Williams, 2007; Steininger et al., 2015). To resolve this issue, we tested propylene glycol-based antifreeze as an alternative to ethanol for preserving benthic arthropod DNA and determined that antifreeze suitably preserves benthic samples for the downstream analysis of macroinvertebrate DNA (Robinson et al., 2020). The application of antifreeze as a preservative in conjunction with training and engaging local communities in remote regions, will enable us to collect samples from areas either only accessible by air or areas where alcohol is prohibited/not available, thus increasing the spatial extent of sample collection (Robinson et al., 2020). For a holistic assessment of freshwater health, it is often necessary to collect data on additional extant taxonomic groups to macroinvertebrates, such as microscopic protist group diatoms (Blanco and Bécares, 2010). The main problem with sampling diatom taxa is the time-consuming and labour-intensive periphyton sampling protocol, which is impracticable for community groups to undertake (Aloi, 1990; King et al., 2006). In order to combine diatom sampling with standard CABIN macroinvertebrate sampling, we tested the taxonomic detection of diatoms from CABIN kick-net sample methodology compared to conventional periphyton microhabitat scraping (Maitland et al., 2020). Through this optimisation, we determined that highly similar diatom assemblages can be detected using solely benthic kick-net sampling, which opens up an important opportunity to simultaneously collect benthos samples for both macroinvertebrate and diatom DNA (Maitland et al., 2020). These two exemplar studies showcases potential for facilitating both remote and multi-taxa sampling for STREAM, highlights the cutting-edge and innovative aspect of the project.
Data processing and reportingHigh-quality data from community-based biomonitoring is possible when standardized field-collection and DNA-based protocols are coupled with high throughput sequencing processed with standardized bioinformatic pipelines (Biggs et al., 2015; Sutherland et al., 2015). STREAM uses high-performing, optimised DNA extraction and sequencing methodologies to generate maximum species coverage for macroinvertebrate community assessment (Hajibabaei et al., 2019). For assessing how effective community-based DNA biomonitoring can be as a model for freshwater monitoring, it is vital to have laboratory protocols optimised fully, with established pipelines and workflow, so that data is processed efficiently and in a timely manner. Existing morphology-based biomonitoring protocols depend on manual morphological taxonomic identifications, which can take longer to generate results and these results are often not presented in an accessible format for non-specialists (i.e. with common names; Fore et al., 2001; Dickinson et al., 2010; Crall et al., 2013). Through applying Biomonitoring 2.0, STREAM will significantly reduce the processing time of samples from up to 12 months or more to less than two months. Generating results within a short time frame is vital for encouraging continued participation from community groups and stakeholders (Baird and Hajibabaei, 2012; Jackson et al., 2016; Keck et al., 2017; Leese et al., 2018).
Using standardized bioinformatic pipelines to process large amounts of sequence data (e.g. (Porter and Hajibabaei, 2018), is essential for scalable, reproducible results that can be directly compared across samples and sites. Pipelines, such as MetaWorks (Porter and Hajibabaei, 2020a), enables data to be analysed in bulk (i.e. 96 samples per sequencing run) and later split into custom DNA reports for community groups and stakeholders. STREAM data reports consist of broad introductory information on biomonitoring and the goals of the project, a succinct methods section and data outputs, including species richness plots and taxa lists which highlight present bioindicator groups. These reports are fully customisable, enabling community groups and stakeholders to include their own study objectives and request additional analyses to meet their objectives. All data, in the form of quality-filtered taxa lists (99% correct classification at family, genus and species level; (Porter and Hajibabaei, 2020a)) with common names in addition to Latin, are sent directly back to community groups and stakeholders along with a custom report. Data, in the form of DNA sequences, is also required to be submitted to open access sequence read archives (SRA), such as on the National Centre for Biotechnology Information SRA (NCBI; https://www.ncbi.nlm.nih.gov/sra), to meet funding agency requirements. To encourage multi-year sampling and to help address long-term biomonitoring objectives, sequences from previous years are re-processed through newer pipeline versions when groups collect samples from the same site(s) over multiple years. This facilitates multi-year comparisons at no additional cost. In addition to physical reports, there is also the option for dissemination of data via virtual meetings, where key information and implications of data generated are fed back to groups.
STREAM data implementationData presented in STREAM reports has been implemented within both community-led and stakeholder environmental programs. For example, independent environmental consulting service, Integrated Ecological Research (IER), partnered with STREAM and a local environmental stewardship group (Slocan River Streamkeepers) to collect and generate multi-year DNA-based monitoring data. This data is being used to assess macroinvertebrate richness and diversity metrics prior to and post-restoration of Slocan River wetlands in British Columbia. In addition, other stakeholders including Parks Canada, Nature Conservancy Canada, 4 Rivers Environmental Services and Fisheries and Oceans Canada (DFO) are currently partnered with STREAM to address biomonitoring objectives across a wide range of watersheds in Canada.
Implementing as nationwide, community-based biomonitoring project such as STREAM, requires effective cross-collaborative partnership. It is important to combine specialities of partners who engage and train community groups, partners who manage databases and provide standardised protocols and lead the project through an organization that will process and handle the community data, to foster communication and meet community (and data) needs. Engaging governmental bodies, such as Environment and Climate Change Canada, within community-based biomonitoring projects is pivotal for routine adoption of DNA-based methods, especially referring to in-kind support and funding (Nerbonne and Nelson, 2004), which is required to address long-term questions concerning freshwater health. Fundamentally, effective communication to community groups and stakeholders is an essential component to success of projects such as STREAM. Communication in the form of resources, contact information, data processing updates and opportunity to provide participation feedback have all proved to be indispensable for creating strong, authentic relationships with community groups.
ConclusionsThe combination of DNA metabarcoding and community-based monitoring is a paradigm shift in our ability to obtain biodiversity information at any time and any place. The information captured could inform existing biomonitoring programmes such as CABIN mainly through taxon-based biodiversity information (e.g. richness and distribution of species or genera of interest). However, molecular analysis provides additional data possibilities that could become highly valuable for next generation of ecological and environmental analysis. By using sequence-based biodiversity measures (e.g. Exact Sequence Variants) one can dramatically increase the information content of samples obtained (Porter and Hajibabaei, 2020b) and inform ecological and environmental models. Furthermore, by using additional DNA markers and more comprehensive sequencing analysis (Singer et al., 2019) it is now possible to monitor biodiversity and trophic relationships of all living organisms in an ecosystem from microbes to mammals.
The uniqueness of STREAM lies in its alliance between academic, non-governmental, government and community groups to carry out freshwater biomonitoring to answer fundamental questions regarding the health of Canada's rivers. This alliance takes advantage of the cutting-edge science as well as people's power in participating in scaling up the application of science within an established regulatory framework for biomonitoring. STREAM sets the stage for how community-based monitoring projects could be applied to global freshwater systems for rapid assessments of biodiversity assemblages to inform ecological analysis. A systematic training design, such as the one used in STREAM, is simple yet effective and can easily be applied to other freshwater systems around the world, if endorsed by NGOs and/or local conservation bodies. The combination of partnering with interested community groups, standardised training and sampling protocols, rapid next-generation sequencing technologies and accessible data packages, emphasises the strength of the STREAM project and the capability of this model to be easily applied outside of Canada.
Authors’ contributionC.V.R. and M.H. conceived the paper, C.V.R. and M.H. wrote the manuscript with input from other co-authors.
This study is funded by the Government of Canada through Genome Canada, Ontario Genomics, and Environment and Climate Change Canada. We would like to thank the community groups and organisations which have partnered with STREAM to enable the co-creation of data.